16S/ITS sequencing and analysis services with expert bioinformatics and statistics.
Start from experimental design, sequencing, or already sequenced data
Reliable, rigorous, and citable results produced with the best available bioinformatics techniques and software
Standard (16S/ITS) and custom amplicons supported
Services
DNA extraction from environmental and clinical samples
Amplicon sequencing: 16S rRNA, ITS, 18S, and other amplicons
Superior Bioinformatics
Comprehensive Report
Interactive Statistical Analysis with ERGO 2.0, includes:
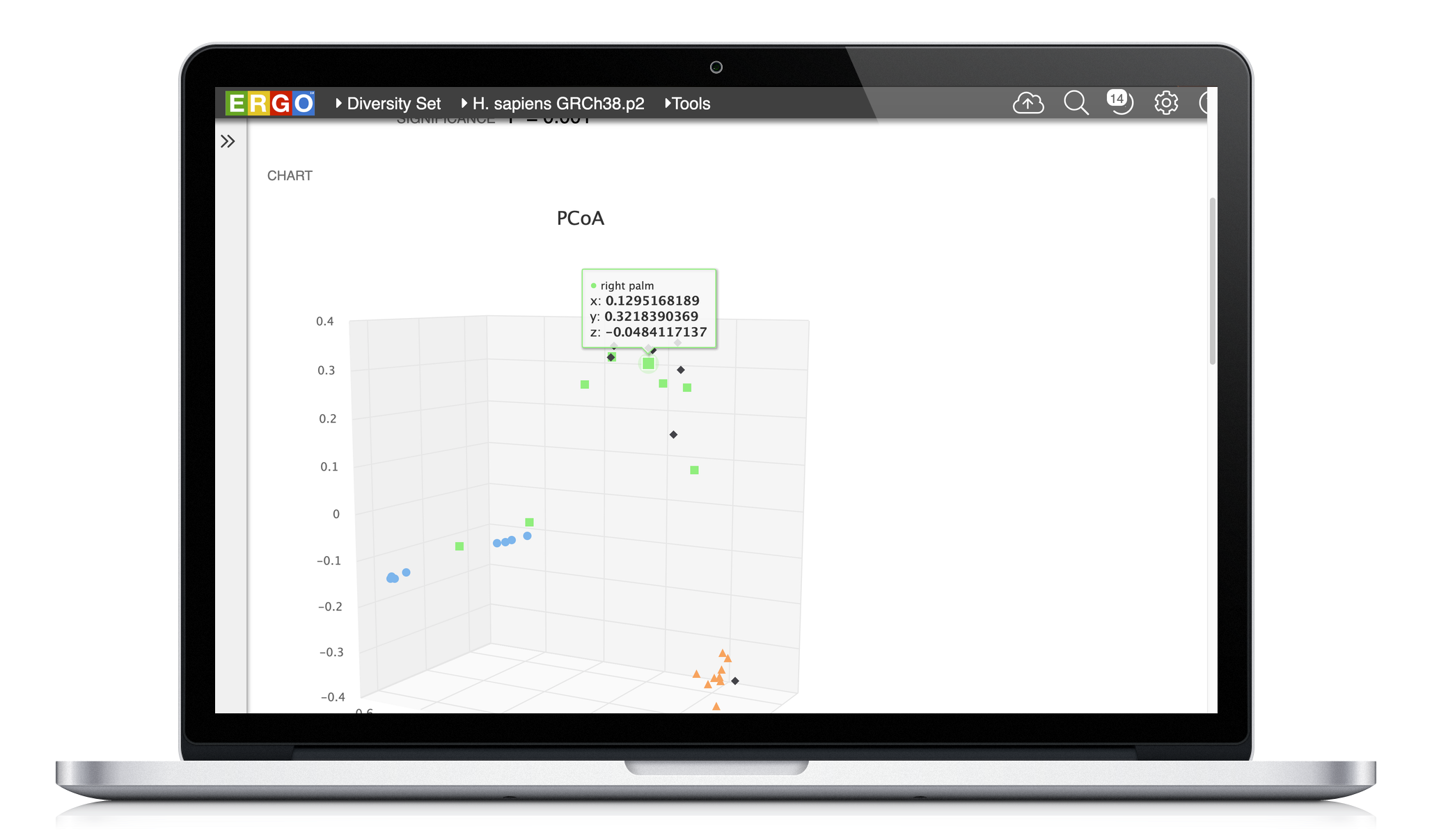
Principal Coordinates Analysis (PCoA)
Analysis of Similarity (ANOSIM)
Linear Discriminate Effect Analysis (for detecting differential abundance)
Alpha-diversity/Richness
Functional Prediction
… And much more, Learn More About ERGO ▸
Applications
Disease and treatment research
Clinical Samples such as skin & cheek swabs, fecal samples, and more.
Environmental Samples such as Soil, River, Sewage, and more.
Industrial samples such as large-scale and desktop fermenters
Identification of Bacteria, Fungi, and other organisms.
Speak with an expert scientist today
Our expert scientists will assist you at every step from experimental design to choosing the right sequencing technology, statistical analysis and visualizations utilizing our best practices microbial community profiling pipelines.
Please complete the form below
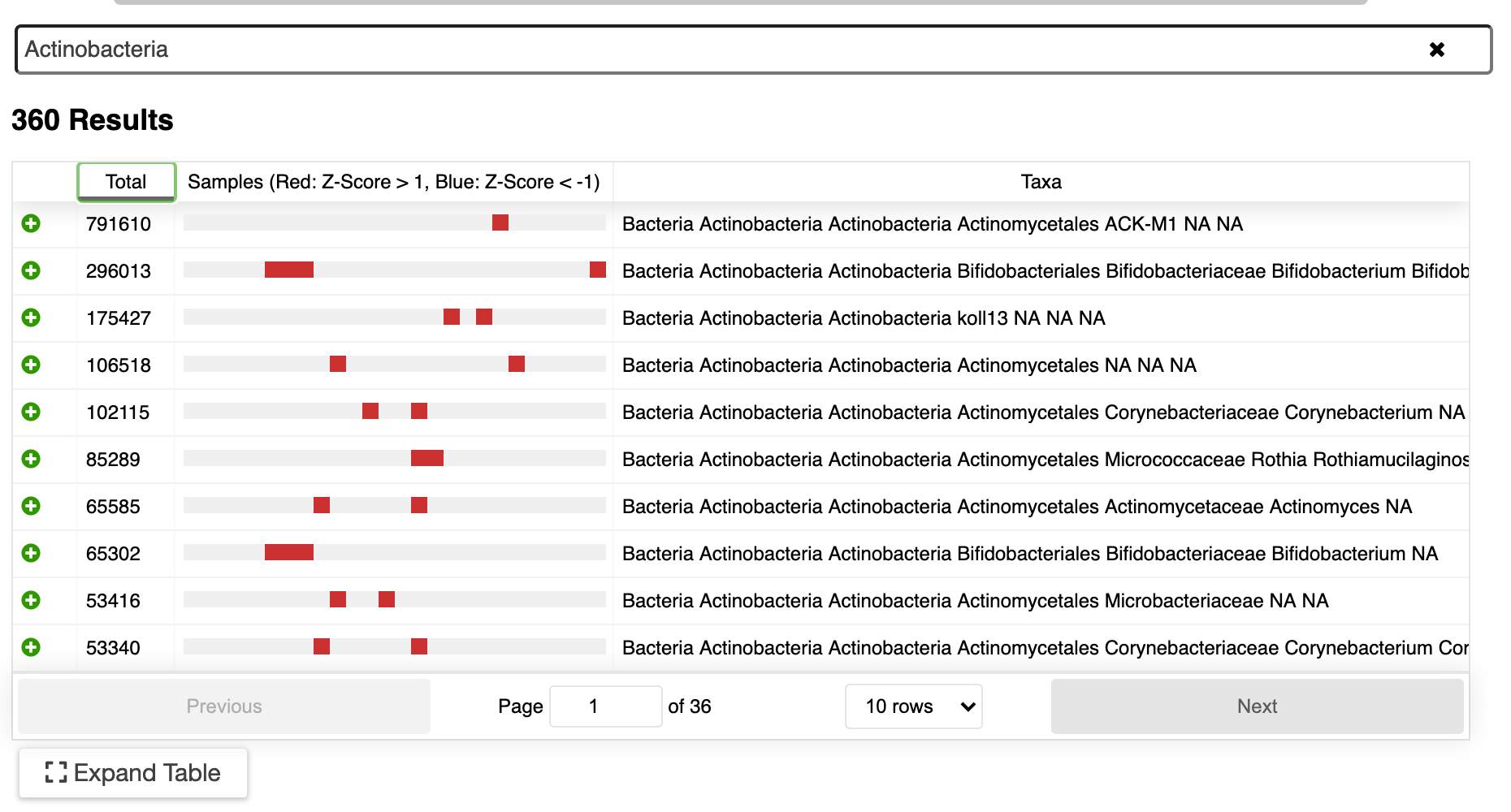
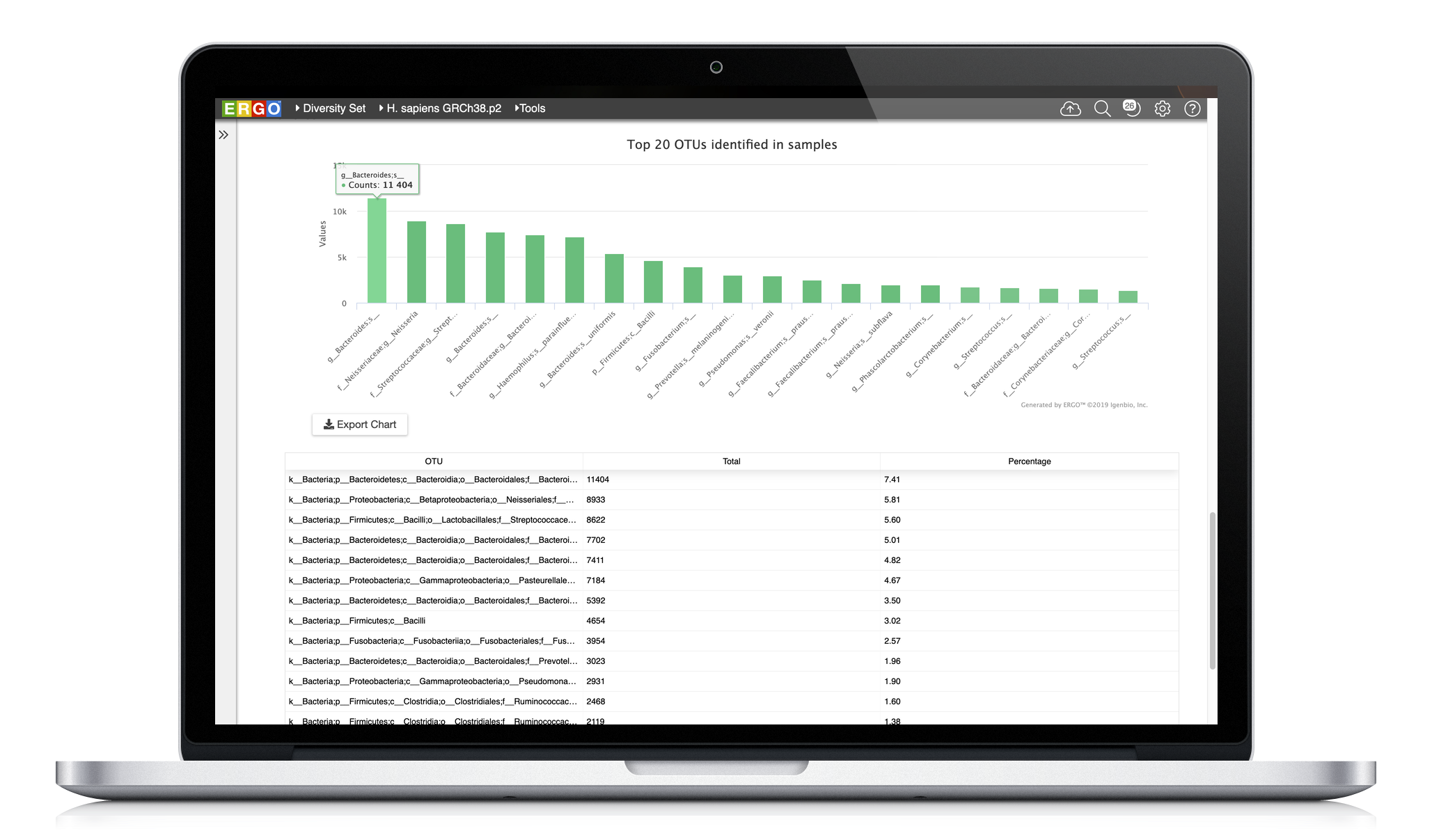
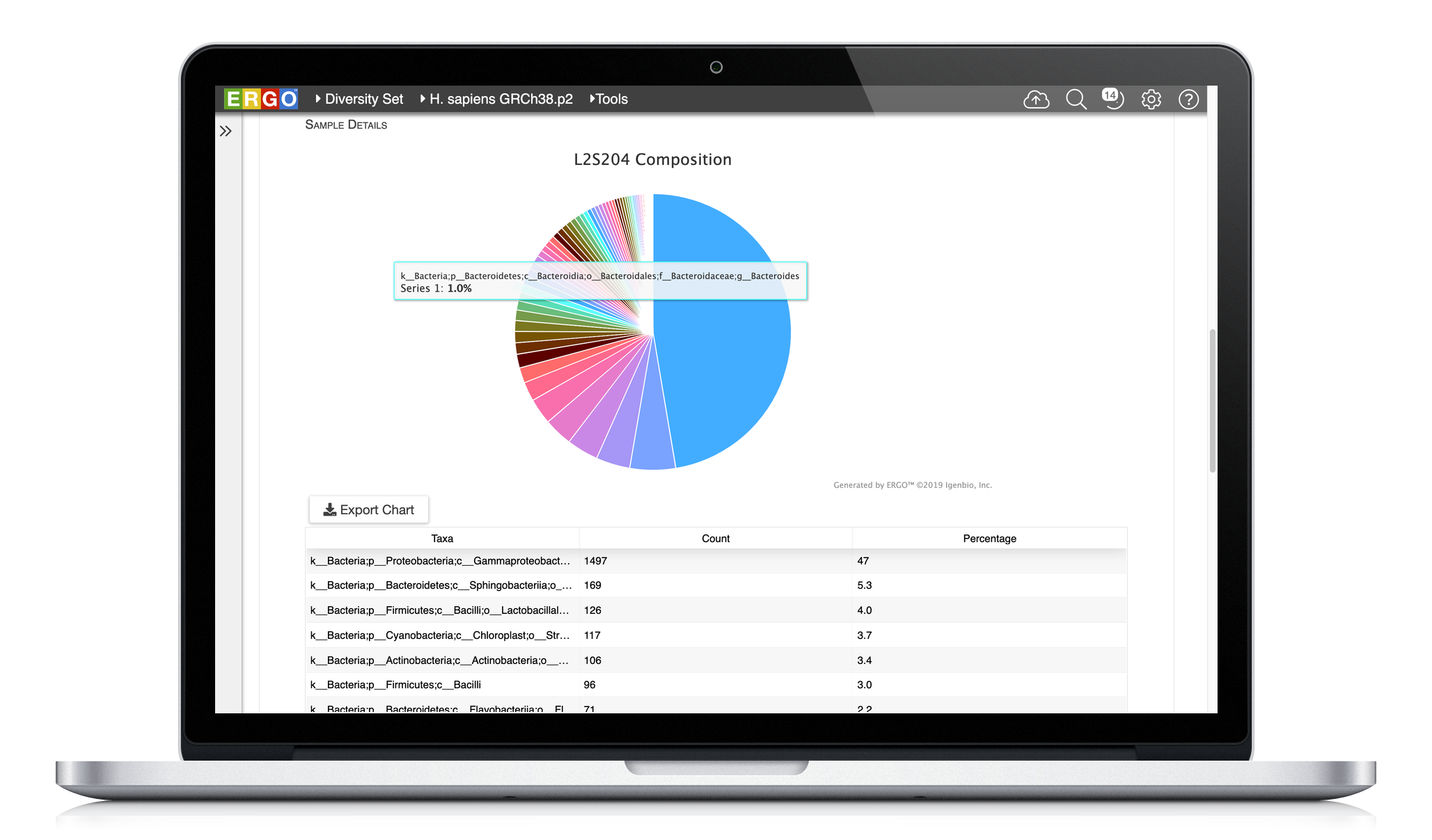
Explore your data with interactive visualizations, including taxonomic assignments, diversity indices, and more.

Why you should choose Igenbio
Proven Track Record
Igenbio, Inc. develops genome analysis products and services for the life science industry. Our scientists have broad experience in both in silico and wet lab sequencing, research and development with more than 100 relevant publications in these areas. Igenbio has a proven track record of delivering scientific results for academic, governmental, and industrial institutions for two decades.
Benefits
Fast 10 business-day Turnaround
Exceptional Sequencing Quality
Sequenced & Analyzed in the United States
Expert support from Igenbio scientists from pellet to plot
Full bioinformatics support available
Interactively explore your data with our informatics suite, ERGO. Learn More ▸
Publications featuring Igenbio Scientists
Related News and Information
Several different software packages such as DADA2, Deblur, and UNOISE use an error-correction technique that is applied to the sequenced amplicon (usually 16S, 18S, or ITS) reads. This ‘denoising’ step is used to increase correct taxonomic assignment of the sequenced amplicons.
We’ve added to ERGO’s rich suite of tools and workflows for 16S amplicon sequencing studies to include functional metagenome prediction utilizing Picrust2.
Do the differences you see on the ordination plot represent a significant difference? This is one of the key questions researchers ask themselves. One way to determine significance is permutational multivariate analysis of variance (PERMANOVA), a statistical test commonly used in ecology settings [1]. ERGO uses the 'adonis' function in the R package vegan which provides functions for the analysis of ecology [2].
Non-metric dimensional scaling attempts to closely represent pairwise dissimilarity between samples. It is a robust unconstrained ordination method that uses rank orders commonly used in ecology studies. Unlike many other ordination techniques NMDS iterates to find a solution that fits with an optimal stress value to the number of chosen dimensions. ERGO gives you a simple interface to choose the number of attempts, dimensions, along with an option to specify a previous best stress.
Data mining and visualization just got easier in ERGO’s newest microbiome analysis update.
ERGO’s [Overbeek et. all 2003] rich suite of tools and workflows to examine amplicon and metagenome sequencing now includes Linear Discriminate Analysis using LEfSe [Segata et. al 2010]. In ERGO, LEfSe (Linear discriminate analysis Effect Size) enables researchers to discover biomarkers that most likely explain the differences between selected experimental groups.
Easily analyze your amplicon sequencing with ERGO’s new drag-and-drop automated Workflow for amplicon sequence analysis. ERGO does it all from sequence read quality checks, primer trimming, denoising, merging, and taxonomic assignment. No knowledge of linux or other tools are necessary.
Our customers commonly ask us what the best primers are to use for an amplicon sequencing project. This article is intended to be a resource to them based on the latest research and our experience performing analysis on numerous projects.
16S rRNA Amplicon Sequencing Primers [2,3,4]
The 16S rRNA small subunit (SSU) is a gene that is part of the 30S subunit of a prokaryotic ribosome. It is often used as a marker since every prokaryotic organism must have a ribosome but the sequence of the 16S component has several regions of variability which are useful for distinguishing between them.
Easily analyze your metagenome projects using ERGO by dragging and dropping your BIOM file into ERGO.